HIGHLIGHTS
Data Management
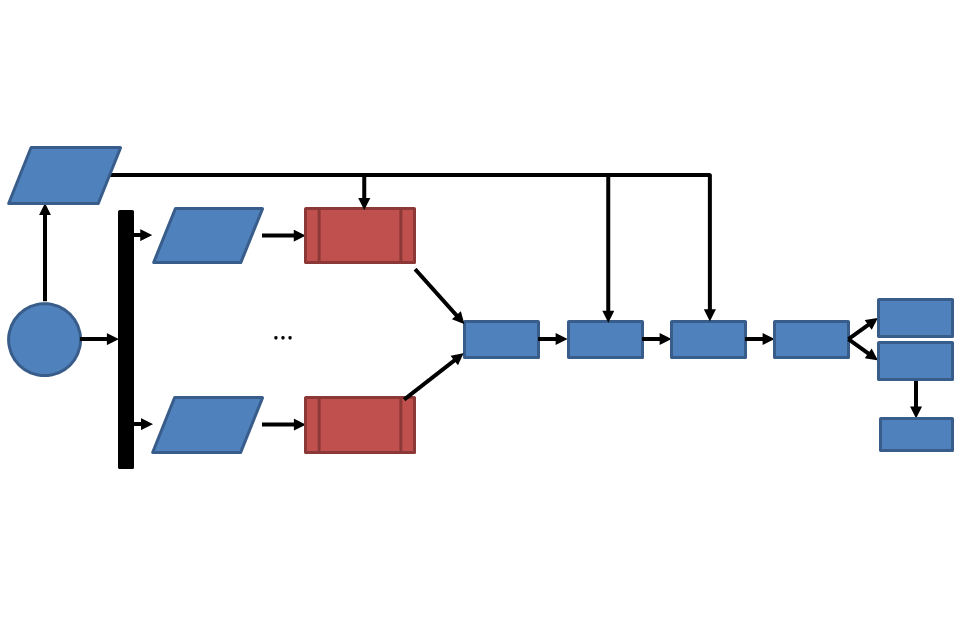
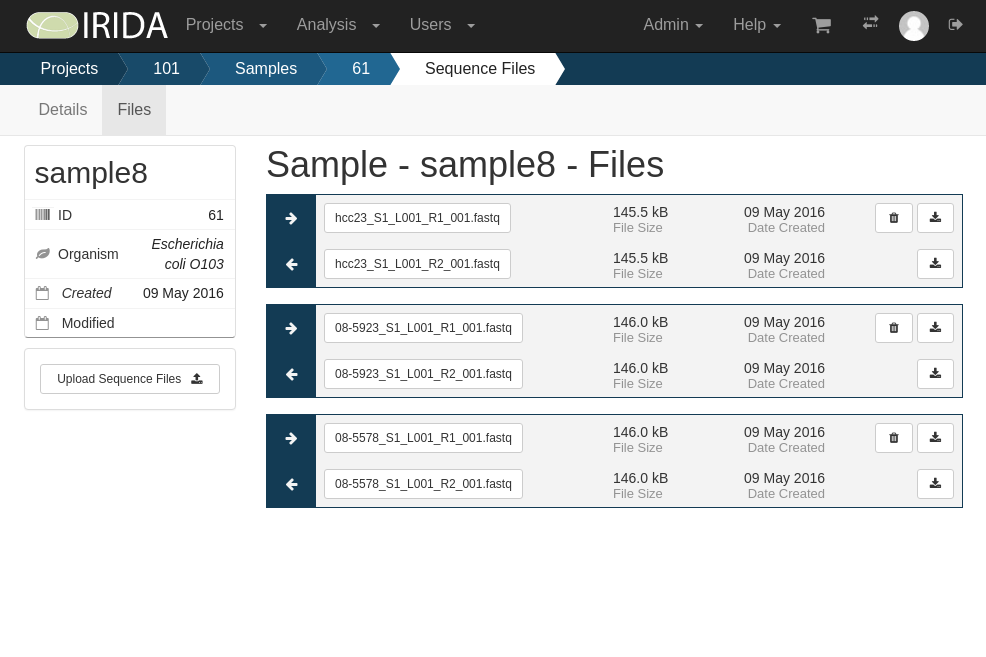
IRIDA provides sequencing data and metadata management functionality to keep track of all your data from sequencer to result.

Data Integration
IRIDA's genomic epidemiology ontology supports the integration of genomics data with epidemiological, lab and clinical information.
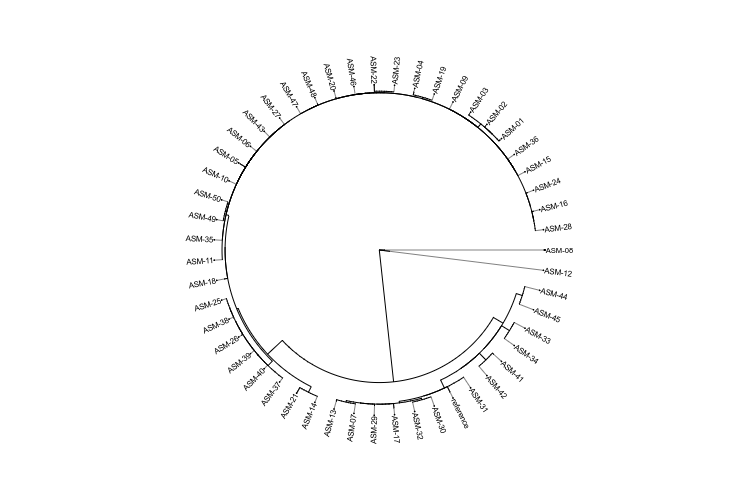
Visualization and Tools
IRIDA contains in-browser data visualization and a REST API for connecting external visualization systems.
INTEGRATED RAPID INFECTIOUS DISEASE ANALYSIS – IRIDA
Public health, food safety, and clinical microbiology labs around the world are embracing whole genome sequencing and genomic epidemiological approaches to modernize their infectious disease research, surveillance, diagnostics, and outbreak investigation programs. This new technology requires powerful, yet easy-to-use, intuitive software in order to transition these new Big Data methods out of the lab and into the front lines of public health activity. IRIDA is designed to make infectious disease genomics accessible to epidemiologists, clinical microbiologists, and the broader research community.
Whole-genome sequencing has proven itself to be an effective epidemiological tool (2008 Listeriosis outbreak, 2009 H1N1 influenza pandemic, 2010 Haiti Cholera outbreak). Unfortunately, the utility of genomic epidemiology to researchers and public health workers is inhibited by the ad-hoc nature and complexity of the bioinformatics software used in genomic epidemiology studies. The primary goal of the IRIDA Bioinformatics Platform is to implement a software platform that makes genomic epidemiology a realistic and usable tool for public health workers, including epidemiologists, clinical microbiologists and researchers.
IRIDA is developed with generous support from and in collaboration with the following agencies and institutions:

LATEST NEWS

Oct 13, 2025
In addition to the active public health version, Simon Fraser University has been hosting a public instance of IRIDA for demonstration purposes. Please note that this public service is fully...

Aug 09, 2025
In addition to the active public health version, Simon Fraser University has been hosting a public instance of IRIDA, currently for demonstration purposes. Please note that this public service is...